Researchers at the University of Chicago Medicine have found new potential drug targets to treat atherosclerosis and coronary artery disease by developing and implementing a new single-cell sequencing workflow that could improve upon those currently in use. The approach will make it easier for other scientists to analyze single-cell genetic data to better understand coronary artery disease as well as other disease states and biological systems.
The team used their cardioinformatics pipeline to reveal new types of vascular cells derived from the smooth muscle cells found in the walls of arteries, along with insight on the communication pathways among those cells.
“This is one of the first studies to show how big data, single-cell sequencing and genomics can be leveraged to find new drug targets in heart disease, which is the number one global killer,” said Bohdan Khomtchouk, PhD, co-senior author on the study and an instructor in the Section of Computational Biomedicine and Biomedical Data Science at UChicago Medicine’s Institute for Genomics and Systems Biology and UChicago’s Master of Science in Biomedical Informatics (MScBMI) program.
Clint Miller, PhD, an assistant professor at the University of Virginia’s Center for Public Health Genomics and study co-senior author, said that the majority of currently approved drugs for heart disease target traditional risk factors such as high cholesterol or blood pressure.
“We hope that these single-cell genomics approaches can inform more precise treatments by targeting cell-specific factors intrinsic to the vessel wall,” said Miller.
Their collaborative study, “Enhanced single-cell RNA-seq workflow reveals coronary artery disease cellular cross-talk and candidate drug targets,” was published November 26, 2021 in the journal Atherosclerosis.
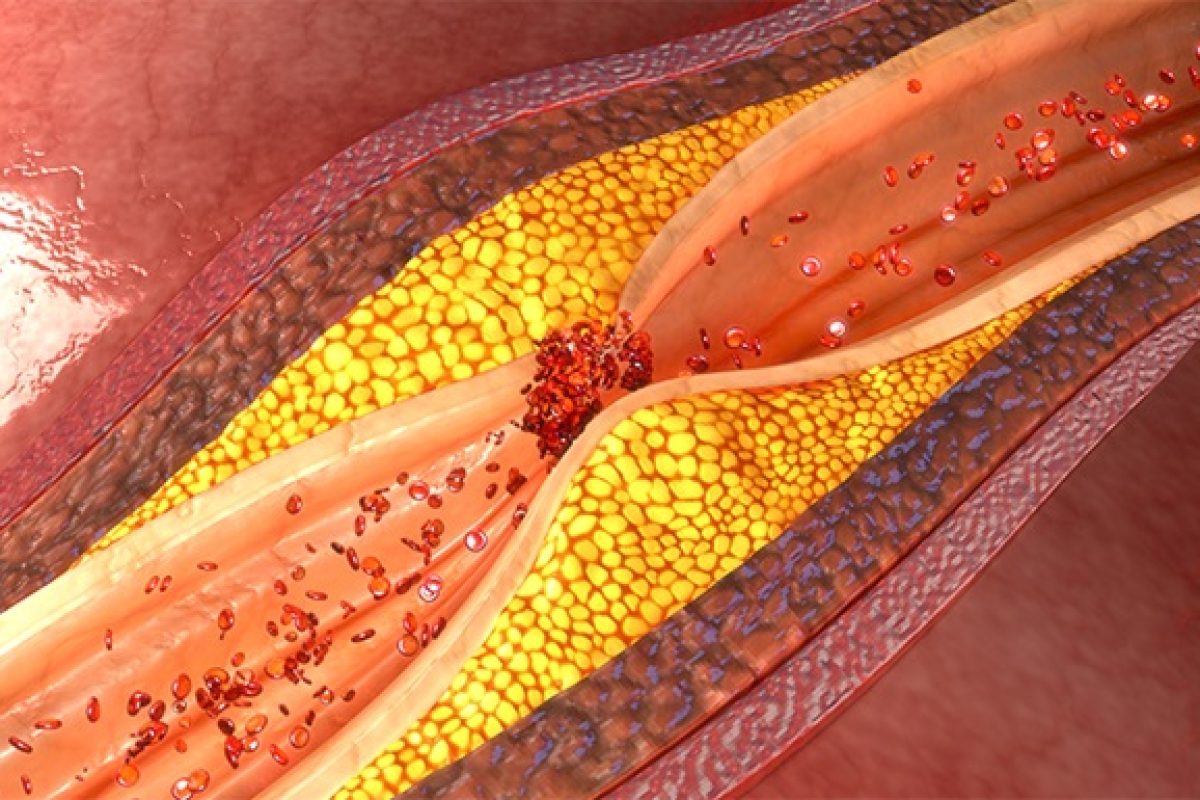
Atherosclerosis is the term given to the build-up of plaque inside or along the walls of arteries, a process that increases gradually as people age. When the vessel walls become inflamed and hardened by the plaque, which is made up of fatty materials like cholesterol, blood flow can be constricted or blocked. Atherosclerosis is a major cause of life-threatening heart attacks and strokes.
Using a modeling technique called RNA-trajectories, the team found that vascular smooth muscle cells give rise to other types of cells, including cells similar to fibroblasts and chondrocytes, that are capable of expressing signs of inflammation and extracellular matrix degradation, both warning signs of atherosclerosis. Khomtchouk likened these vascular cell discoveries to finding new clusters of stars in the sky.
“Scientists are still discovering different cell types in the heart, and this is where we broke new ground,” said Khomtchouk, whose lab specializes in using computational biology and large, complex sets of biological data to research and discover new drugs for cardiovascular, renal and metabolic diseases. “We wanted to show that we can find new heart cell types – cells that don’t even have names yet – using some of these large-scale cardioinformatics approaches.”
The team was able to identify several already-approved drugs as capable of disrupting communication between those smooth muscle cells and fibroblast cells. For instance, they identified several chemotherapies targeting the epidermal growth factor receptor (EGFR) signaling cascade that hold potential for drug repurposing to block erroneous signaling and inflammation in different cell types.
Understanding how this “crosstalk” among cells works could help scientists better understand the complexities of atherosclerosis and how plaque begins to develop; the next step would be verifying the effectiveness of the drugs by conducting preclinical and clinical trials.
While the research is a preliminary first step to developing a new drug, these types of computer-performed in silico experiments shed data-driven light on potential combination therapies or reduce potential adverse effects by targeting cell type-specific processes in the heart and vasculature.
“Over 90 percent of clinical trials fail,” said Khomtchouk. “Any kind of precision cardiology effort that can make drug discovery more precise is extremely valuable and informs actionable analytic insights that illuminate novel cardiovascular biology and enable new therapeutics.”
The workflow has applications beyond atherosclerosis and coronary artery disease research. Khomtchouk and Miller hope that scientists in different fields such as cancer and neurodegenerative diseases apply the workflow to their own datasets to formulate testable biological hypotheses.
The researchers also developed an open-source, interactive web application called PlaqView that allows anyone to explore datasets and compare single-cell RNA sequencing tools with no prior coding knowledge.
“More human atherosclerosis and coronary artery disease datasets are actively being added to PlaqView, including disease contexts from other species like mouse,” said Khomtchouk. “More data is more statistical power. Our lab is pioneering the field of cardioinformatics and studies like this are paving the road towards a healthier, more sustainable future of better heart drugs through computation.”
“Enhanced single-cell RNA-seq workflow reveals coronary artery disease cellular cross-talk and candidate drug targets” was written by Bohdan Khomtchouk and Alexandra Ligay of the University of Chicago, Clint Miller, Wei Feng Ma, Chani Hodonsky, Adam Turner, Doris Wong, Yipei Song, Jose Verdezoto Mosquera, Christina Gancayco, Gary Owens, Nelson Barrientos, David Mai, Gabriel Alencar and Katherine Owsiany of the University of Virginia, Lotte Slenders, Gerard Pasterkamp, Michal Mokry and Sander van der Laan of Utrecht University and Huize Pan and Muredach Reilly of Columbia University. Funding support was provided by the National Institutes of Health, the American Heart Association and the Leducq Foundation Transatlantic Network of Excellence.
Originally published at UChicagoMedicine.org